gst_prime
Implementation of a standardized genetic differentiation measure
G′ST
The R code presented here calculates the G′ST estimator used in:
Tumonggor MK, TM Karafet, B Hallmark, JS Lansing, H Sudoyo, MF Hammer and MP Cox. 2013. The Indonesian archipelago: an ancient genetic highway linking Asia and the Pacific. Journal of Human Genetics 58: 165-173.
Tumonggor MK, TM Karafet, S Downey, JS Lansing, P Norquest, H Sudoyo, MF Hammer and MP Cox. 2014. Isolation, contact and social behavior shaped genetic diversity in West Timor. Journal of Human Genetics 59: 494-503.
The G′ST estimator was developed by:
Hedrick PW. 2005. A standardized genetic differentiation measure. Evolution 59: 1633-1638.
Specifically, the code implements equation 4b in Hedrick (2005):
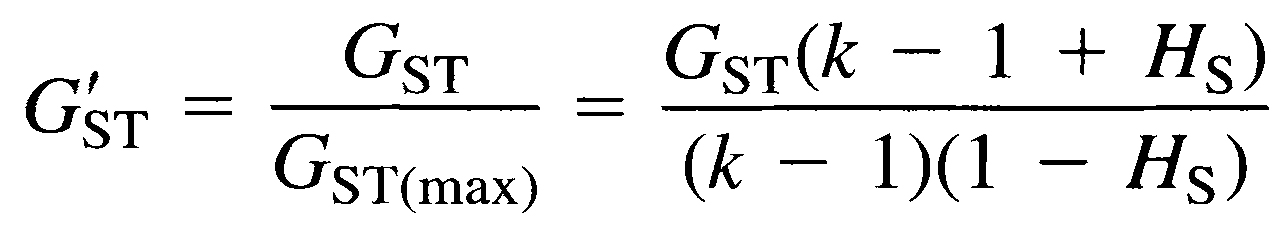
The gst_prime function is written in base R, and requires values for the number of subpopulations k, the average subpopulation heterozygosity HS and the total population heterozygosity HT. Usage is:
gst_prime(k, HS, HT)
Worked example:
A study population containing 14 subpopulations with average subpopulation heterozygosity of 0.953 and total population heterozygosity of 0.981 would return:
gst_prime(14, 0.953, 0.981)
0.6518016